Recherche translationnelle en santé,
technologie pour la santé et recherche clinique
Stéphane Chavanas’ research is oriented towards the pathological modelling and physiopathology of human diseases.
Previous works
At INSERM U385 unit in Nice, then at the Wellcome Trust Centre for Human Genetics in Oxford, Stéphane Chavanas studied the biology and pathophysiology of the epidermis, and characterised genetic mutations causing severe skin diseases, such as the mutations within the gene SPINK5 that he discovered (e.g. : *1–4 (see refernces).
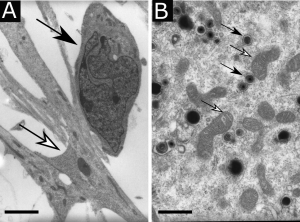
Transmission electron microscopy showing (A) a human neural stem cell (white arrow) in contact with an immature neuron (black arrow), scale bar : 5 µm, or (B) cytomegalovirus viral particles (white arrows) in the cytoplasm of a neural stem cell in the vicinity of mitochondria (black arrows), scale bar : 0,5 µm.
At Toulouse Purpan Physiopathology Centre (now Infinity, Toulouse Institute for Infectious and Inflammatory Diseases), Stéphane Chavanas studied the pathophysiology of congenital infection by human cytomegalovirus, which is the most common cause of congenital abnormalities of the central nervous system. In collaboration with the Institut des Cellules Souches ISTEM (Corbeil-Essonnes), Stéphane Chavanas has developed an in vitro model of HCMV infection of neural progenitors with neural stem cells derived from human embryonic stem cells (Fig. 1). The studies of Stéphane Chavanas’ group have provided with a better knowledge of the molecular and cellular mechanisms underlying the sequelae of infection in placenta *5 or in fetal brain *6,*7.
Human brain organoids
Production of brain organoids from human pluripotent cells was first achieved in 2013 by M. Lancaster and J. Knoblich. It was a key step towards modelling healthy or pathological human brain. During maturation, these organoids lead to the formation of brain-like structures and the differentiation of progenitors into a variety of neuronal or glial cell types. However, these organoids, which are grown in suspension, do not faithfully reproduce brain anatomy and raise issues regarding reproducibility and nutrients and oxygen supply to all cell populations. As part of the new 3D Chip team, we are developing an original 3D bioprinted- human brain organoid model.
We will use human neural stem cells (NSC) derived from embryonic stem cells (Fig. 2 and 3). We will also use genetically modified NSCs or NSCs produced from iPSCs (induced pluripotent cells) to model diseases of the central nervous system (autism, neurodegenerative diseases, etc.). The cells will be encapsulated in hydrogels to optimise their proliferation and differentiation. The 3D bioprinting technology will provide precision, reproducibility and flexibility. These models could reproduce the diversity and organisation of the brain at the cell level (progenitors, neurons, macroglia and microglia…) They will be characterised by microscopy with specific markers, by electrophysiology and by magnetic resonance imaging. Our models will have a strong potential for pathological modelling and physiopathology studies or for drug screening.armaceutiques.
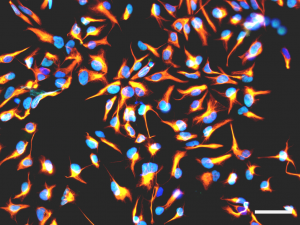
Figure 2 : Immunofluorescence analysis showing cultured human neural stem cells expressing nestin (orange) or SOX2 (cyan). Nuclei are counterstained with DAPI (blue). Scale bar : 50 µm.
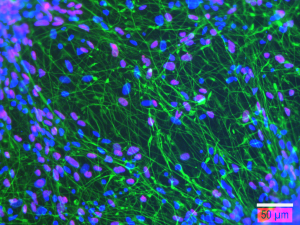
Figure 3 : Immunofluorescence analysis showing human neural stem cell-derived neurons after 21 days of differentiation in vitro and expressing beta 3 tubulin (green), or HUC/D (magenta). Nuclei are counterstained with DAPI (blue). Scale bar : 50 µm.
References
*1 : Chavanas, S. et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat. Genet. 25, (2000).
*2 : Walley, A. J. et al. Gene polymorphism in Netherton and common atopic disease. Nat. Genet. 29, (2001).
*3 : Chavanas, S. et al. A homozygous nonsense mutation in the PLEC1 gene in patients with epidermolysis bullosa simplex with muscular dystrophy. J. Clin. Invest. 98, (1996).
*4 : Chavanas, S. et al. Splicing modulation of integrin ?4 pre-mRNA carrying a branch point mutation underlies epidermolysis bullosa with pyloric atresia undergoing spontaneous amelioration with ageing. Hum. Mol. Genet. 8, (1999).
*5 : Leghmar, K. et al. Cytomegalovirus Infection Triggers the Secretion of the PPARgamma Agonists 15-Hydroxyeicosatetraenoic Acid (15-HETE) and 13-Hydroxyoctadecadienoic Acid (13-HODE) in Human Cytotrophoblasts and Placental Cultures. PLoS One 10, e0132627 (2015).
*6 : Rolland, M. et al. PPARγ Is Activated during Congenital Cytomegalovirus Infection and Inhibits Neuronogenesis from Human Neural Stem Cells. PLoS Pathog. 12, e1005547 (2016).
*7 : Rolland, M. et al. Human cytomegalovirus infection is associated with increased expression of the lissencephaly gene PAFAH1B1 encoding LIS1 in neural stem cells and congenitally infected brains. J. Path. 254:92-102 (2021)

